StochDynTools
Status: Work in progress. The documentation is fairly complete, but this software
is still in its earlier stages of development and perhaps not very user
friendly. Please email me if you have questions/problems using it. I’ll be
happy to help.
Requires the symbolic toolbox (MuPAD version, which is now the default) and all tests were done with MATLAB R2010a.
Download: [tar file for MuPAD symbolic toolbox -- used by
R2010a and later] (recommended)
[tar
file for MAPLE symbolic toolbox -- used by R2008a] (not recommended)
This set of MATLAB scripts provides a set of functions that compute approximate moment dynamics for a stochastic network of chemical reactions. This approximation is based on the moment closure techniques described in the following references:
J. Hespanha. Moment closure for biochemical networks. In Proc. of the Third Int. Symp. on Control, Communications and Signal Processing, Mar. 2008. [pdf]
J. Hespanha, Abhyudai Singh. Stochastic Models for Chemically Reacting Systems Using Polynomial Stochastic Hybrid Systems. Int. J. on Robust Control, Special Issue on Control at Small Scales: Issue 1, 15(15):669—689, Sep. 2005.
A. Singh, J. Hespanha. LogNormal Moment Closures for Biochemical Reactions. In Proc. of the 45th Conf. on Decision and Contr., Dec. 2006.
A typical set of chemical reactions is specified by a .net file of the following form:
% Filename:
enzyme_with_Et_St.che
% Michaelis-Menten
kinetics, keeping track of one fast variable (free enzyme)
% and three slow variables
(total subtrate, total enzymes, product)
species:
EF stochastic 2; %
free enzyme (fast
variable)
Et deterministic 3; % total enzymes = free + compound (slow variable)
St stochastic 11; %
total substrate = free + compound (slow variable)
P
stochastic 2; % product (slow variable)
parameters:
K_C =
1; % rate constant
for S + EF -> ES
D_C = 20; % rate constant for ES -> S + EF
K_P =.05; % rate constant for ES -> P + EF
reactions:
% fast reversible reaction
rate = K_C*(St-Et+EF)*EF; {EF} > {EF-1}; % S + EF -> ES
rate = D_C*(Et-EF); {EF} > {EF+1}; % ES -> S + EF
% slow product reaction
rate = K_P*(Et-EF); {St,P,EF} > {St-1,P+1,EF+1}; % ES
-> P + EF
% END of .net file
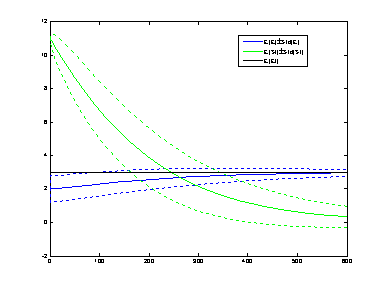
The following commands produce the evolution of the moment dynamics shown in the figure below
net=readNet('enzyme_with_Et_St.net');
mdyn2=closureDynamics(net,2,'derMatch','fun');
Tmax=600; % maximum simulation time
[t,mu]=ode23s(@(t,x)fun(x),[0,Tmax],mdyn2.Mu.x0);
h=plotCMoments(net,mdyn2,t,mu,'EF','b-','St','g-','Et','k-');
legend(h(1:3:end),'E[EF]\pm{}Std[EF]','E[St]\pm{}Std[St]','E[Et]','location','best');
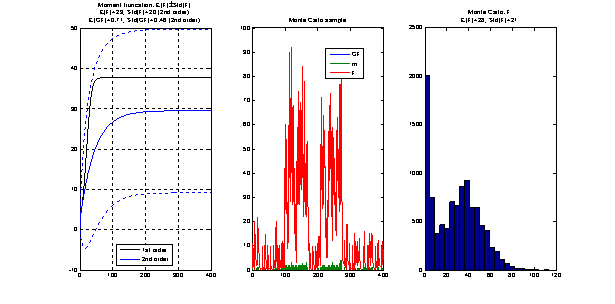
The following picture is
taken from an example included in the package and compares the moments dynamics
obtained by truncation with Monte Carlo simulations.
Bibliographic citation
When used in research, please acknowledge the use of this software with the following reference:
João Hespanha. StochDynTools — a MATLAB toolbox to compute moment dynamics for stochastic networks of bio-chemical reactions. Available at http://www.ece.ucsb.edu/~hespanha, May. 2007.
or if you use latex/bibtex:
@Misc{HespanhaDec06,
key =
{matlab,c,biology},
author =
{João Pedro Hespanha},
title =
{\texttt{StochDynTools} --- a {MATLAB} toolbox to
compute moment dynamics for
stochastic networks of
bio-chemical reactions},
howpublished = {Available at
\url{http://www.ece.ucsb.edu/~hespanha}},
month =
may,
year =
2007
}
Disclaimer
This program
is free software; you can redistribute it and/or modify it under the terms of
the GNU General Public License as published by the Free Software Foundation;
either version 2 of the License, or (at your option) any later version. This
program is distributed in the hope that it will be useful, but WITHOUT ANY
WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A
PARTICULAR PURPOSE. See the GNU General
Public License for more details (http://www.gnu.org/copyleft/gpl.html)