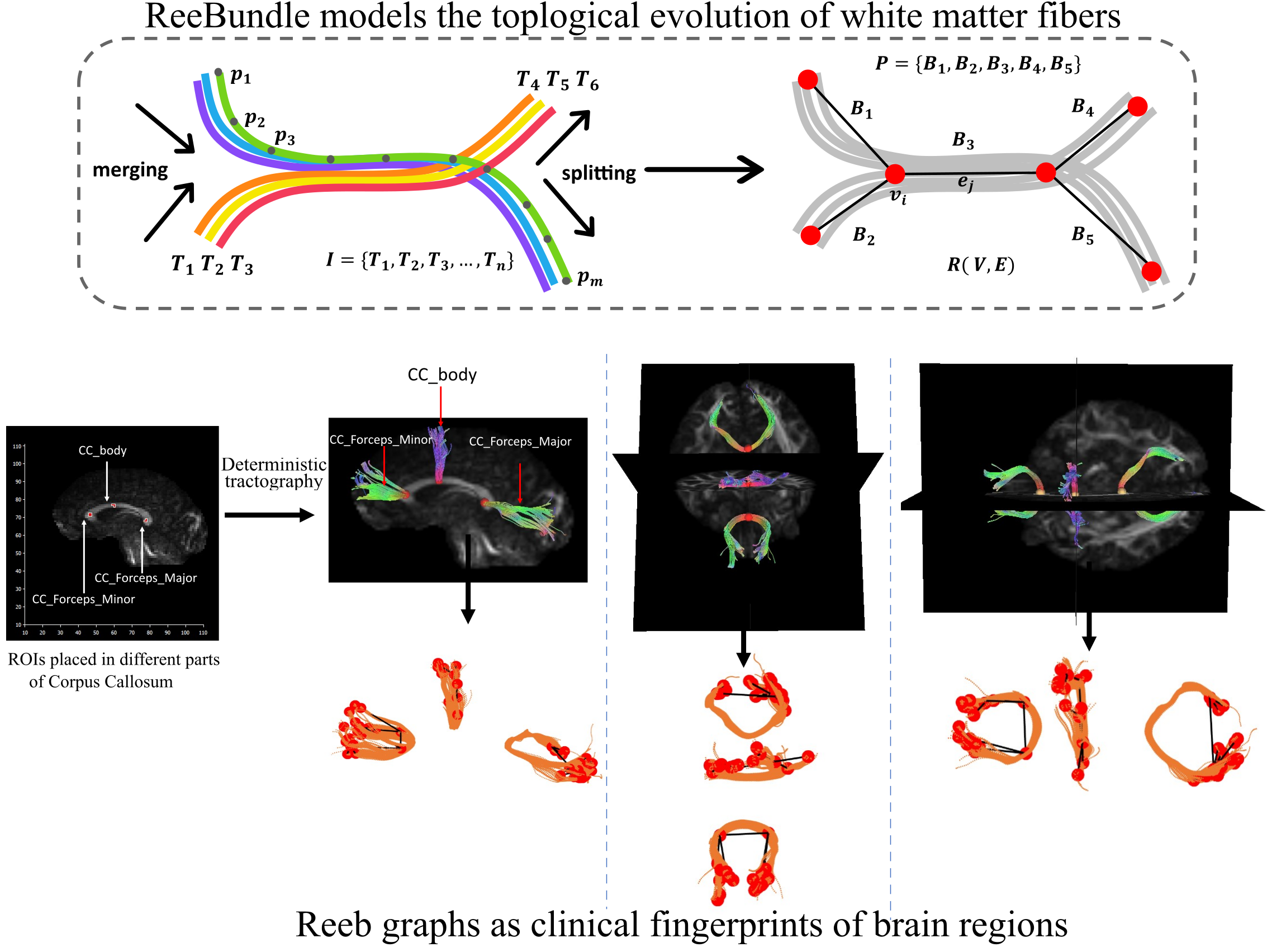
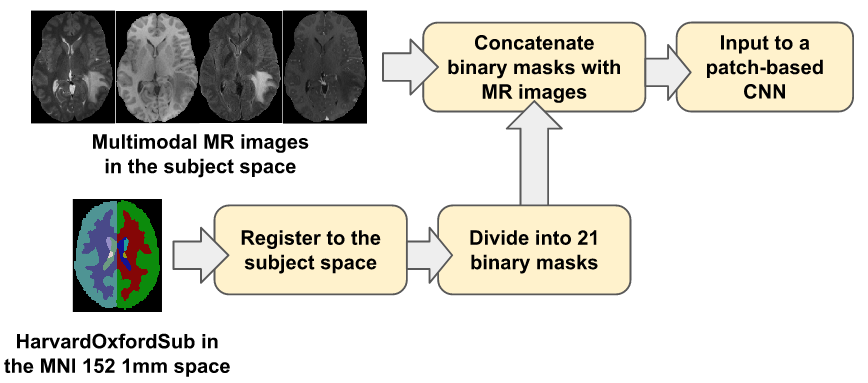
Brain Connectivity Analysis
Reeb Graph for brain connectome
Our brain is a complicated anatomical network responsible for cognition and behavior. Neurodegenerative diseases impede the associated network architecture in millions of affected people. My research aims to mathematically model the neuronal activities in the human brain using tools from computational geometry and artificial intelligence to characterize the evolution of neuronal fibers. We model the high-level topological structures of the fibers from diffusion MRI imaging data to construct graph-based mathematical objects. My research will pave the way for advanced AI methods to compare brain regions and provide pathological insights that are currently infeasible due to the complexity of the neuronal fibers.